HLA Antigen Level Matching
The primary role of the HLA molecules is to present pathogen derived peptides to T cells thereby eliciting a T cell mediated adaptive immune response. The T cell recognises both the HLA molecule and the peptide it presents, distinguishing self derived peptides from foreign peptides. It is this ability to restrict the T cell response, distinguishing self from foreign and permitting an immune response to be mounted against the foreign that makes the HLA antigens the main immunological barrier to transplantation, necessitating HLA matching.
The first successful living-related donor kidney transplant was performed in 1954 by a team in Boston, USA and involved a kidney transplant between 23-year-old identical twins. These identical twins were matched at all 6 classical HLA loci and the patient survived for many years. Many studies have since shown a strong correlation between the level of HLA matching at the Broad level for HLA-A, B and DR and graft survival. In the UK in the 1990’s, it was shown that the best outcome was achieved with kidneys that had no mismatches at HLA-A, HLA-B, and HLA-DR loci (000 mismatches). The next most favourable outcome was achieved with one mismatch at either A or B loci or one mismatch at both the A and B , but no mismatch at the DR locus (100, 010, or 110 mismatches).
A recent UNOS study has suggested a reduction of the influence of HLA matching in kidney transplantation in the Calcineurin Inhibitor era and with improvements in surgical techniques. This has however been refuted by a European CTS study (Opelz et al) which reviewed transplants over two decades from 1985 to 2004. The European study found a statistically significant association between graft survival and the number of mismatches across that period. They also found a significant association between the number of mismatches and the number of rejection episodes.
In the UK an updated HLA matching algorithm was implemented in 2006. The previous matching scheme was potentially iniquitous to patients from ethnic minorities, with potentially rarer HLA types, who are under represented on the donor panels. A system of ‘defaulting’ of rare HLA antigens to common equivalents was introduced. e.g. HLA-A80 in a patient was defaulted to HLA-A1 and HLA-DR103 was defaulted to HLA-DR1, making it much more likely that such patients will be transplanted.
The current UK scheme comprises 5 tiers as far as HLA matching is concerned. Tiers A comprises 000 matched highly sensitised paediatric patients and 000 matched HLA-DR homozygous paediatric patients. Tier B comprises all other 000 matched paediatric patients. Tier C comprises 000 matched highly sensitised adult patients and 000 matched HLA-DR homozygous adult patients. Tier D comprises of all other 000 matched adult patients and all well matched paediatric patients (100, 010, and 110 HLA-A, -B, and -DR mismatches). All other patients are in Tier E. A point system is then used to prioritise within each tier based on factors including waiting times.
The emphasis that the scheme places on prioritising 000 matched paediatrics highlights the impact of mismatching of HLA antigens on the development of alloantibodies and therefore the ability to re-transplant in later years should the graft fail. The data the current UK algorithm is based on four identified levels (graded levels 1 – 4) of HLA matching which were found in a review of transplant outcomes based on the UK 1998 algorithm, to correlate well with increasing risk of transplant failure. Level 1 comprises 000 HLA-A, B and DR mismatches, Level 2 comprises 0 HLA-DR and 0 or 1 HLA-B mismatch, Level 3 comprises 0 HLA-DR and 2 HLA-B mismatches or 1 HLA-DR and 1 HLA-B mismatch and Level 4 comprises 1 HLA-DR and 2 HLA-B mismatches or 2 HLA-DR mismatches. The UK data showed that HLA-A mismatches had no effect on transplant outcome.
The impact of matching for HLA-C, DQ and DP in kidney transplantation has been reviewed in a small number of studies. One study has suggested a potential influence of HLA-C mismatching on the number of acute rejection episodes in the presence of 1 additional HLA-B mismatch. The role of HLA-DQ mismatching in the presence of a HLA-DR match has received relatively little study in recent years. One study in the 1980’s and another in the 1990’s found no significant correlation between HLA-DQ mismatching and graft outcome in the presence of a HLA-DR match. A small number of studies have shown a role for HLA-DP matching in re-transplant patients but no significant role in first transplants even in the presence in preformed donor specific antibodies.
HLA matching for kidney transplantation is generally at the Broad antigen level and not at the allele level. One study has shown a correlation between allele level mismatched in HLA-DRB1 and the number of rejection episodes though the study found no correlation with long term survival. Certainly the ability to identify allele specific antibodies using the current generation of solid phase HLA antibody detection techniques presents the ability to list allele specific antibodies as unacceptable mismatches. In addition, a number of studies have shown that the use of structural epitope matching techniques such as HLAMatchmaker is predictive of positive crossmatches. Duquesnoy has shown that in 0 HLA-DR matched, HLA-A and/or HLA-B mismatched transplants, the number of epitope mismatches correlates significantly with 5 year graft survival.
The impact of HLA matching in kidney transplantation continues to evolve in the desensitisation era with the presence of preformed donor specific alloantibodies no longer the absolute contraindication to transplantation that it once was. The UK BSHI/BTS guidelines require that laboratories are capable of identifying HLA antibodies to HLA-A, B, C, DR, DQ and DP so that donors who should be negative can be identified for crossmatching. However desensitisation protocols may permit transplantation even in the presence of donor specific antibodies for any of these HLA loci.
Not all solid organ transplantations require the level of HLA matching that is the norm in kidney transplantation though most require HLA antibody definition. Pancreatic transplantation is one exception which does requires the same level of HLA matching as kidney transplantation. With cardiothoracic transplantation, HLA matching is not necessarily undertaken but antibody definition is required and if present then a prospective crossmatch or retrospective crossmatch within 48 hours is required.
HLA matching prior to liver transplantation is not required and prospective crossmatching is not indicated. Some centres do carry out HLA antibody identification to aid in setting immunosuppression levels. HLA matching is not necessarily undertaken for small bowel and intestinal transplantation.
Epitope Matching
It has long been known that antibodies do not bind to the whole of the HLA antigen but instead bind to specific epitopes on the antigen surface. Each HLA antigen potentially has many sites or epitopes that can bind antibody. These epitopes may be private to a given HLA antigen or they may be shared by more than one HLA antigen, i.e. they may be public. Several studies have demonstrated a potential application to solid organ, especially renal transplantation.
Several epitope systems have been described, including the Terasaki system which is based on use of Luminex SAB to absorb and elute samples and identify the specific epitope the antibody is reacting with, the Duquesnoy HLAMatchmaker in silico system and the Kosmoliaptsiset system which is based on interlocus and intralocus comparison of patients and donors to identify amino acid differences but also crucially, the physiochemical properties of the amino acid mismatches and the role these may play in clinical outcome.
There are two versions of HLAMatchmaker, the Triplet and Eplet versions. The Triplet version treats HLA class I antigens as linear sequences of sequences of 3 amino acids in antibody accessible regions of the HLA molecule. Amino acids in positions such as the β pleated sheet are considered not to be in antibody accessible regions and are specifically excluded. The Eplet version is based on an analysis of the three dimensional structure of the HLA molecule rather than on linear sequences of amino acids. Instead of epitopes being defined by three amino acids in a linear sequence, eplets are defined as all the amino acids within a 3 to 3.5Å radius of each polymorphic residue position.
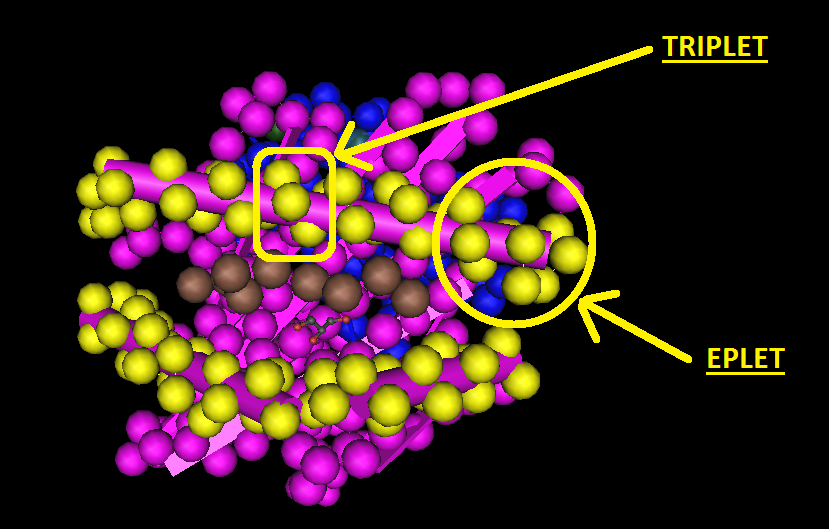
The applicability of epitope matching to kidney transplantation, particularly using HLA Matchmaker, has been extensively studied. In the antibody testing and crossmatching setting, studies have shown that a higher number of epitope mismatches between donor and patient has a statistically significant association with presence of Luminex positive HLA antibodies and with a positive crossmatch. In one large study, Duquesnoy et al, showed that in 0DR mismatched kidney transplants, 5 years outcomes for transplants with kidneys that had up to 4 Triplet mismatches, irrespective of the number of antigen mismatches, had the same organ survival rates as those patients transplanted with no HLA-A or B antigen mismatches.
Another study showed that matching at the epitope level rather than the antigen level, potentially increased the number of donors that could be considered compatible for a sensitised patient.
The number of epitope mismatches between patient and donor has also been shown to be statistically associated with the frequency of de novo donor specific antibody formation post-transplant.